
PMET
Find Co-Occurrences of TF Binding Site Motifs on Sequences
Why Choose PMET?
PMET is a powerful tool designed to assist researchers in identifying the interactions of transcription factors (TFs) to regulate gene network. By studying the combinations of homotypic and heterotypic motifs within transcription regulatory modules, PMET provides a comprehensive framework for analyzing and understanding the functional implications and regulatory dynamics associated with motif interactions in gene expression.
PMET is designed to address the limitations of traditional analysis tools by considering both homotypic and heterotypic motif combinations simultaneously.
PMET is available in both command-line and web-based versions, providing flexibility and convenience to researchers.
How to Use PMET
Essentially all you need to run PMET for your biological process of interest is a set of genomic regulatory sequences. These could be promoters of differentially expressed genes, differentially open chromatin regions, regions identified from DNA methylation assay, or regions identified using other techniques.
Run job
1. Select PMET running mode: PMET provides three running modes depending on your mode of retrieval of sequences.
2. Upload target sequences with clusters: Upload sequences ID chosen in specific clusters for regulatory.
3. Set parameters: Adjust documented PMET parameters or use the default settings.
After completing the steps above, you can initiate PMET for analysis.
Visualization
PMET also offers visualization tools to analyze the distribution of paired motifs on genes. It is often found that genes in different clusters exhibit enrichment for specific motif-pairs.
Functionality of PMET
Homotypic Clustering: PMET can identify clusters of homotypic motifs within the genome based on the motif data provided by the user. This analysis helps uncover the significance and functionality of motifs in gene regulation.
Heterotypic Clustering: After identifying clusters of homotypic motifs,PMET further analyzes the pairings between these clusters to generate heterotypic clusters. Through this process, PMET reveals the potential interactions between motifs in gene regulation.
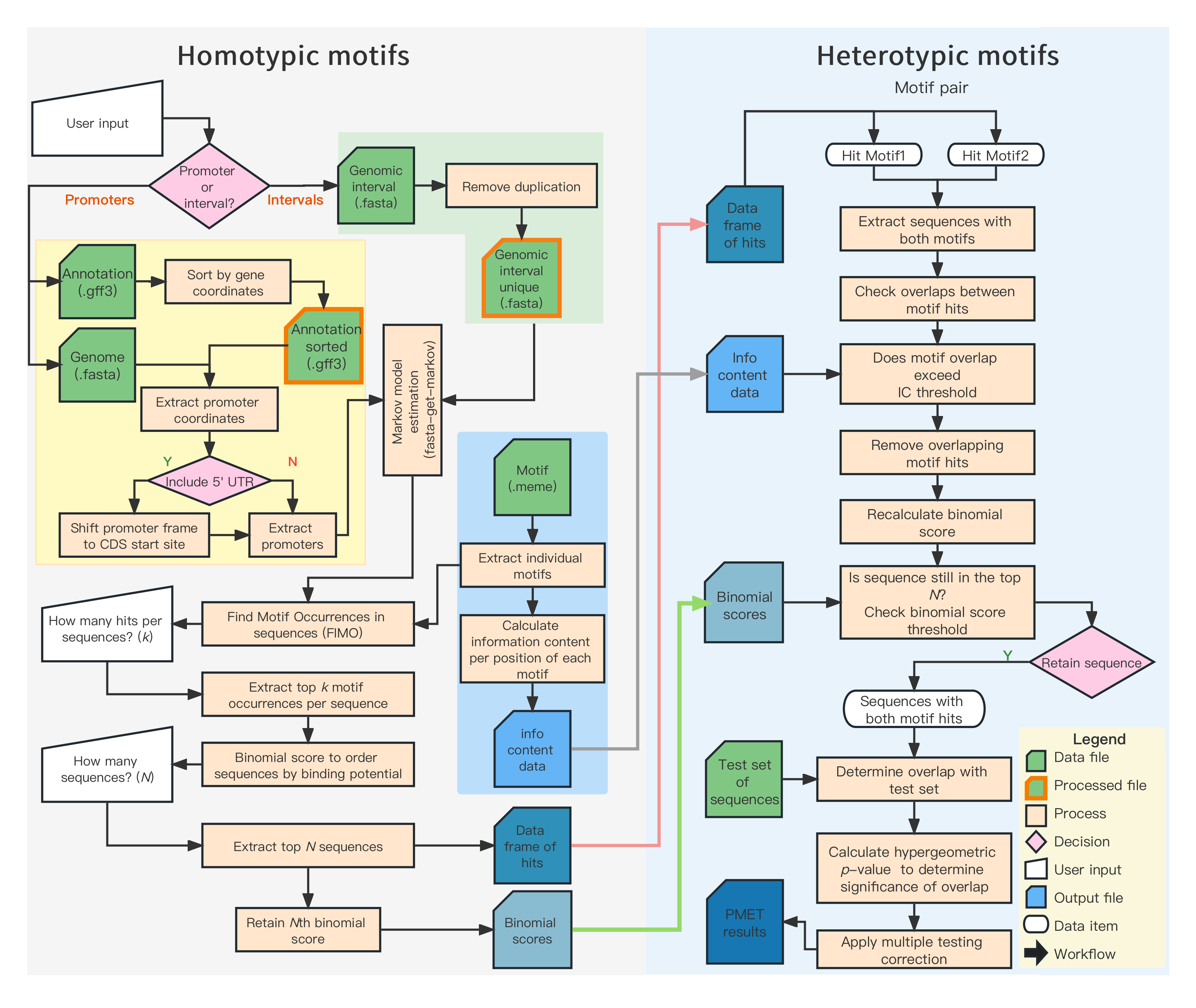
Workflow of PMET computation
e-mail: s.ott@warwick.ac.uk
email: Patrick.Schaefer@agrar.uni-giessen.de
e-mail: k.woolley-allen@warwick.ac.uk
e-mail: p.e.brown@warwick.ac.uk
e-mail: Xuesong.Wang@agrar.uni-giessen.de